pan-european assessment, monitoring, and mitigation of stressors on the health of bees
Validated qPCR methods open perspectives in nosemosis diagnosis and bee epidemiology
Pathogens and parasites, and their associated diseases, are among the major stressors affecting bee health and consequently pollinator decline. The unicellular eukaryote Nosema microsporidia (Nosematidae) are among some of the widespread parasites of managed and wild bees. Nosema apis and Nosema ceranae have been described in honey bees and Nosema bombi in bumble bees. And although there are molecular methods which serve as a complement to microscopic diagnosis of nosemosis, to date, there are only a few developed methods allowing accurate quantification of these three Nosema species and data on infections by them in the same bee species are scarce.
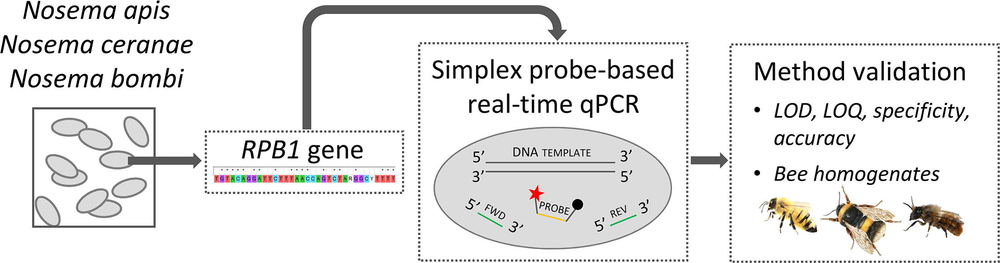
Graphical abstract from the "Specific detection and quantification of three microsporidia infecting bees, Nosema apis, Nosema ceranae, and Nosema bombi, using probe-based real-time PCR"
A recent PoshBee article – Specific detection and quantification of three microsporidia infecting bees, Nosema apis, Nosema ceranae, and Nosema bombi, using probe-based real-time PCR – describes three quantitative real-time PCRs (qPCRs) for the independent detection and quantification of the three Nosema species in honey bees, bumble bees, and mason bees. The paper also reports validation parameters, which include PCR efficiency, specificity, limit of detection, limits of quantification and quantification accuracy.
These qPCR methods open perspectives in nosemosis diagnosis and in bee epidemiology. To find out more about them, read the full paper here.